(modified last February 05, 2008)
Although ~95% of deletions can be detected in males using multiplex PCR, other methods must be used to determine duplications, as well as the carrier status of females. The most commonly applied methods are quantitative multiplex PCR and quantitative Southern blotting. The drawback of quantitative multiplex PCR is that often not all exons are examined, meaning that small and rare mutations are missed. Southern blotting is usually applied to determine the boundaries of the mutation, which is essential for distinguishing DMD from BMD, i.e. frame disrupting from open reading frame changes. Using high-quality Southern blots it is possible to perform a quantitative analysis and detect duplications. However, this technique is time consuming, it is difficult to exactly determine the duplication boundaries, it can be difficult to detect duplications in females and triplications will be missed.
We have developed a method to screen the entire DMD gene for deletions and duplications (White et al. [2002]). This method uses MAPH, Multiplex Amplifiable Probe Hybridisation, and analyses all 79 exons of the DMD gene in one assays (see DMD MAPH analysis). Recently, a similar method has been designed, based on MLPA (Multiplex Ligation-dependent Probe Amplification, Schouten et al. [2002]). The advantage of MLPA compared to MAPH is that a lower amount of input DNA is required and that MLPA is a one-tube assay.
A kit for MLPA analysis of the human DMD-gene can be obtained from MRC-Holland (SALSA P034 / P035 DMD test kit) as well as detailed MLPA-protocols. A paper describing our results using this kit has been published by Lalic et al. (2005). In addition we have published results using a modified kit that allows array-based MLPA-screening of the DMD-gene (Zeng et al. 2008). For those interested we offer deletion / duplication screening of the DMD-gene using both MLPA (for information please contact us; Johan den Dunnen - E-mail: ddunnen @ HumGen.nl).
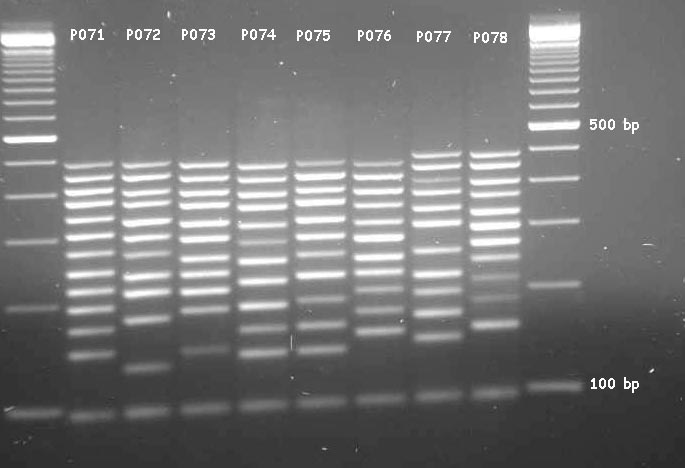
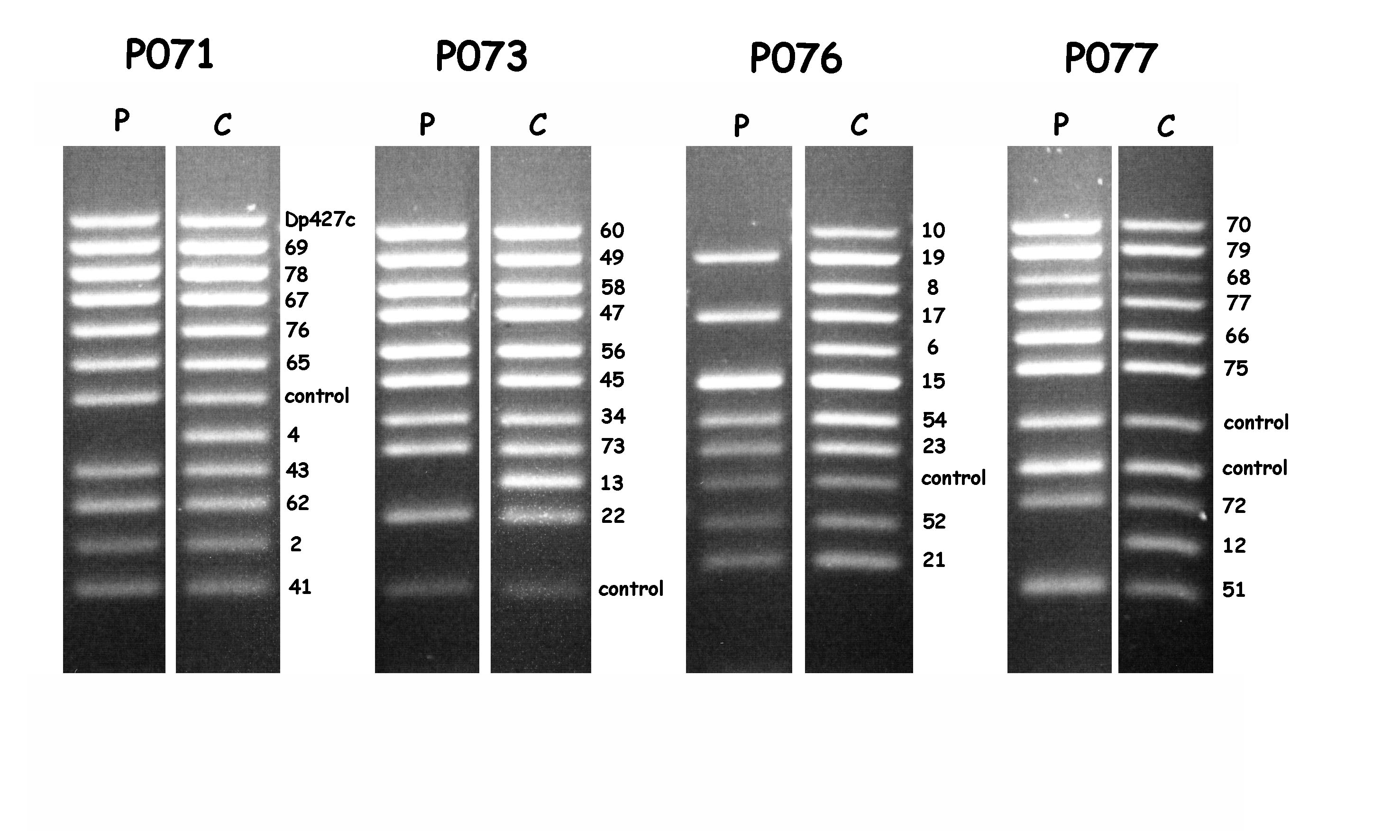
Usually, MLPA-products are fluorescently labelled and analysed using capillary gel-electrophoresis (e.g. using an automated DNA sequencer - see MLPA reference pictures). A nice feature of the MLPA-analysis of the DMD gene is that the amplification products can also be analysed using simple agarose gel-electrophoresis and ethidium-bromide staining. The only thing to do in this respect is to order the DMD MLPA-kit in a specific format. An example of the analysis of the 79-exons of the DMD-gene in this way is shown in Fig.1. An example of the analysis of a DMD-patient carrying a deletion is shown in Fig.2. When the number of PCR-cycles is optimised, agarose gel- electrophoresis even facilitates the detection of duplications in patients and deletions in carriers (White et al., in preparation).

Legend:
MLPA analysis of the human DMD-gene in a normal male. Content and length of the individual products is indicated in Table I.
Picture kindly provided by Stefan White and Rolf Vossen.
Legend:
MLPA-analysis using agarose gel-electrophoresis of a DMD patient with a deletion
of exons 4 to 13. Missing exons can be easily detected using the individual
sets; P071 - exon 4, P073 - exon 13, P076 - exons 6, 8 and 10, P077 - exon 12.
Not all sets are shown. Picture kindly provided by Stefan White and Rolf Vossen.
| P071 | P072 | P073 | P074 | P075 | P076 | P077 | P078 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| probe | bp | probe | bp | probe | bp | probe | bp | probe | bp | probe | bp | probe | bp | probe | bp |
| exon 41 | 146 | control | 130 | control | 142 | exon 11 | 138 | Dp427m | 138 | exon 21 | 154 | exon 51 | 146 | exon 31 | 154 |
| exon 2 | 170 | exon 42 | 178 | exon 22 | 186 | exon 61 | 162 | exon 71 | 162 | exon 52 | 178 | exon 12 | 170 | control | 184 |
| exon 62 | 194 | exon 3 | 210 | exon 13 | 210 | exon 32 | 186 | control | 186 | control | 202 | exon 72 | 194 | control | 211 |
| exon 43 | 218 | exon 63 | 234 | exon 73 | 234 | exon 53 | 218 | exon 33 | 226 | exon 23 | 226 | control | 218 | exon 14 | 242 |
| exon 4 | 242 | exon 74 | 266 | exon 34 | 258 | exon 44 | 250 | exon 24 | 258 | exon 54 | 250 | control | 250 | exon 64 | 266 |
| control | 274 | exon 25 | 298 | exon 45 | 290 | exon 5 | 282 | exon 55 | 290 | exon 15 | 282 | exon 75 | 306 | exon 35 | 298 |
| exon 65 | 306 | exon 36 | 330 | exon 56 | 322 | exon 16 | 314 | exon 46 | 322 | exon 6 | 314 | exon 66 | 338 | exon 26 | 330 |
| exon 76 | 338 | exon 27 | 370 | exon 47 | 362 | exon 7 | 354 | exon 57 | 362 | exon 17 | 354 | exon 77 | 378 | exon 37 | 370 |
| exon 67 | 378 | exon 38 | 402 | exon 58 | 394 | exon 18 | 386 | exon 48 | 394 | exon 8 | 386 | exon 68 | 410 | exon 28 | 402 |
| exon 78 | 410 | exon 29 | 442 | exon 49 | 434 | exon 9 | 426 | exon 59 | 434 | exon 19 | 426 | exon 79 | 450 | exon 39 | 442 |
| exon 69 | 450 | exon 40 | 483 | exon 60 | 475 | exon 10 | 467 | exon 50 | 475 | exon 10 | 467 | exon 70 | 491 | exon 30 | 483 |
| Dp427c | 491 | ||||||||||||||
Legend
Individual DMD MLPA-probe sets P071 to P078 (obtained from MRC-Holland).
Probe: probe for DMD-exon indicated or a control probe. bp: length
of MLPA amplification product in base pairs (bp). An agarose-gel of these
products can be seen in Fig.1.
| Top of page | LMDp homepage | Diagnostic techniques |
| Remarks / information | Disclaimer
|