(last modified March 3, 2007)
Multiplex PCR is a technique in which several PCR-products are amplified in one
PCR-reaction. DMD is an X-linked disease, i.e. only one X-chromosome is present.
Consequently, deletion of an exon can be determined by the failure to amplify
that exon by PCR. Since deletion mutations are frequent in DMD/BMD (present in
~2/3 of cases) and since the deletions are non-randomly distributed across the
gene (see "DMD gene deletion/duplication database"),
a multiplex PCR co-amplifying the most frequently deleted exons provides a very
simple, rapid and efficient diagnostic tool. DMD multiplex PCR was first described by Chamberlain
et al. (1988) and became rapidly applied world-wide. The original
6-exon Chamberlain-set was later modified into a 9-exon (Chamberlain
et al. 1990) and ultimately a 10-exon set (Beggs
et al. 1991).
A second set, the 9-exon Beggs-set (Beggs
et al. 1990),
was developed to increase the total number of deletions detected and to define the borders
of the deletions in the deletion 'hotspot'. To cover some untested regions and
to facilitate the determination of the borders of the deletions in a larger
number of cases, Kunkel et al. 1991 developed a third 7-exon set.
For specific purposes, several additional multiplex primer sets were published
over the years (see below). An extensive discussion of the DMD/BMD multiplex PCR protocol
and its characteristics can be found in Current Protocols in Human
Genetics (by Alan Beggs and Johan den Dunnen).
NOTE: a 10-set quantitative multiplex PCR assay
was descibed by Stockley et al. (2006).
When detection of point mutations in the DMD gene was still rather difficult, several studies used the the multiplex PCR products trying to identify such mutations. The techniques applied include single-strand conformation analysis (SSCA), hetero-duplex analysis (HD) and double-strand conformation analysis (DSCA). With these technologies, frequent polymorphic sequence variants can be detected in:
Please note that since some of the primer sequences used in the multiplex sets are exonic, span or are to close to the exon/intron boundaries, this approach will fail to detect mutations which are at the primer annealing site and those outside the amplified region, in particular those affecting splicing. For efficient methods to scan the DMD gene for point mutations see "DNA-based diagnostic techniques for DMD / BMD".

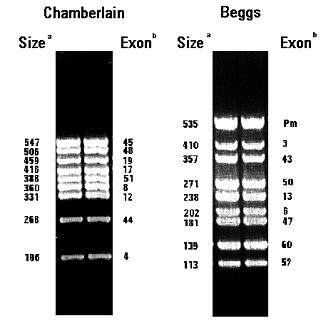
Legend:
Size: size of PCR product (in base pairs). Exon: exon of DMD gene
present in PCR fragment.
(listed in the order the fragments appear on gel)
| Amplified exon | in set | Reference | Length (in bp) |
Forward / reverse primer |
Name |
|---|---|---|---|---|---|
| exon 45 | a | Chamberlain 1990 | 547 | aaacatggaacatccttgtggggac / cattcctattagatctgtcgccctac |
ex45-F/R |
| exon 48 | b | Chamberlain 1990 | 506 | ttgaatacattggttaaatcccaacatg / cctgaataaagtcttccttaccacac |
ex48-F/R |
| exon 19 | c | Chamberlain 1990 | 459 | gatggcaaaagtgttgagaaaaagtc / ttctaccacatcccattttcttcca |
ex19-F/R |
| exon 17 | d | Chamberlain 1990 | 416 | gactttcgatgttgagattactttccc / aagcttgagatgctctcacCTTTTCC |
ex17-F/R |
| exon 51 | e | Chamberlain 1990 | 388 | gaaattggctctttagcttgtgtttc / ggagagtaaagtgattggtggaaaatc |
ex51-F/R |
| exon 8 | f | Chamberlain 1990 | 360 | ggcctcattctcatgttctaattag / gtcctttacacactttacCTGTTGAG |
ex8-F/R |
| exon 12 | g | Chamberlain 1990 | 331 | gatagtgggctttacttacatccttc / gaaagcacgcaacataagatacacct |
ex12-F/R |
| exon 44 | h | Chamberlain 1990 | 268 | cttgatccatatgcttttacctgca / tccatcacccttcagaacctgatct |
ex44-F/R |
| exon 4 | i | Chamberlain 1990 | 196 | ttgtcggtctctctgctggtcagtg / caaagccctcactcaaacatgaagc |
ex4-F/R |
NOTE: the Chamberlain-set was extended with a 10th fragment (fragment J) for exon 46 by Beggs et al. (1991). PCR primers used were GCTAGAAGAACAAAAGAATATCTTGTC (ex46-F) and CTTGACTTGCTCAAGCTTTTCTTTTAG (ex46-R) giving a product of 148 bp.
(listed in the order the fragments appear on gel)
| Amplified exon | in set | Reference | Length (in bp) |
Forward / reverse primer |
Name |
|---|---|---|---|---|---|
| Dp427m exon 1 | a | Beggs 1990 | 535 | GAAGATCtagacagtggatacataacaaatgcatg / ttctccgaaggtaattgcctcccagatctgagtcc |
PmF/R |
| exon 3 | b | Beggs 1990 | 410 | tcatcc a tcatcttcggcagattaab / caggcggtagagtatgccaaatgaaaatca |
ex3-F/R |
| exon 43 | c | Beggs 1990 | 357 | gaacatgtcaaagtcactggacttcatgg / atatatgtgttacctacCCTTGTCGGTCC |
ex43-F/R |
| exon 50 | d | Beggs 1990 | 271 | caccaaatggattaagatgttcatgaat / tctctctcacccagtcatcacttcatag |
ex50-F/R |
| exon 13 | e | Beggs 1990 | 238 | aataggagtacctgagatgtagcagaaat / ctgacCTTAAGTTGTTCTTCCAAAGCAG |
ex13-F/R |
| exon 6 | f | Beggs 1990 | 202 | ccacatgtagGTCAAAAATGTAATGAA / gtctcagtaatcttcttacCTATGACTATGG |
ex6-F/R |
| exon 47 | g | Beggs 1990 | 181 | cgttgttgcatttgtctgtttcagTTAC / gtctaacCTTTATCCACTGGAGATTTG |
ex47-F/R |
| exon 60 | h | Beggs 1990 | 139 | AGGAGAAATTGCGCCTCTGAAAGAGAACG / CTGCAGAAGCTTCCATCTGGTGTTCAGG |
ex60-F/R |
| exon 52 | i | Beggs 1990 | 113 | AATGCAGGATTTGGAACAGAGGCGTCC / TTCGATCCGTAATGATTGTTCTAGCCTC |
ex52-F/R |
NOTE: Sylvie Chambert pointed out that the genomic sequence for this primer differs; tcatccGtcatcttcggcagattaa. Since PCR failure has not been reported, efficient primer annealing seems not to be comprised by this difference.
(listed in the order the fragments appear on gel)
| Amplified exon | in set | Reference | Length | Forward primer | Name |
|---|---|---|---|---|---|
| exon 49 | a | Beggs 1990 | 439 | gtgcccttatgtaccaggcagaaattg / gcaatgactcgttaatagccttaagatc |
ex49-F/R |
| Dp427c exon 1 | b | Kunkel 1991 | 332 | TCTGGCTCATGTGTTTGCTCCGAGGTATAG / CTTCCATGCCAGCTGTTTTTCCTGTCACTC |
Pb-F/R |
| exon 16 | c | Kunkel 1991 | 290 | tctatgcaaatgagcaaatacacgc / ggtatcactaacCTGTGCTGTACTC |
ex16-F/R |
| exon 41 | d | Kunkel 1991 | 274 | gttagctaactgccctgggccctgtattg / tagagtagtagttgcaaacacatacgtgg |
ex41-F/R |
| exon 32 | e | Kunkel 1991 | 253 | gaccagttattgtttgaaaggcaaa / ttgccaccagaaatacatacCACACAATG |
ex32-F/R |
| exon 42 | f | Kunkel 1991 | 195 | CACACTGTCCGTGAAGAAACGATGATGG / CTTCAGAGACTCCTCTTGCTTAAAGAGAT |
ex42-F/R |
| exon 34 | g | Kunkel 1991 | 171 | GTAACAGAAAGAAAGCAACAGTTGGAGAA / CTTTCCCCAGGCAACTTCAGAATCCAAA |
ex34-F/R |
(listed in the order the fragments appear on gel)
| Amplified exon | in set | Reference | Length (in bp) |
Forward / reverse primer |
Name |
|---|---|---|---|---|---|
| Dp427m exon 1 | a | Abbs 1991 | 535 | GAAGATCtagacagtggatacataacaaatgcatg / ttctccgaaggtaattgcctcccagatctgagtcc |
PmF/R |
| exon 19 | b | Abbs 1991 | 459 | gatggcaaaagtgttgagaaaaagtc / ttctaccacatcccattttcttcca |
ex19-F/R |
| exon 3 | c | Abbs 1991 | 410 | tcatccatcatcttcggcagattaa / caggcggtagagtatgccaaatgaaaatca |
ex3-F/R |
| exon 8 | d | Abbs 1991 | 360 | ggcctcattctcatgttctaattag / gtcctttacacactttacCTGTTGAG |
ex8-F/R |
| exon 13 | e | Abbs 1991 | 238 | aataggagtacctgagatgtagcagaaat / ctgacCTTAAGTTGTTCTTCCAAAGCAG |
ex13-F/R |
| exon 6 | f | Abbs 1991 | 202 | ccacatgtagGTCAAAAATGTAATGAA / gtctcagtaatcttcttacCTATGACTATGG |
ex6-F/R |
| exon 4 | g | Abbs 1991 | 196 | ttgtcggtctctctgctggtcagtg / caaagccctcactcaaacatgaagc |
ex4-F/R |
NOTE: Sylvie Chambert pointed out that the genomic sequence for this primer differs; tcatccGtcatcttcggcagattaa. Since PCR failure has not been reported, efficient primer annealing seems not to be comprised by this difference.
(listed in the order the fragments appear on gel)
| Amplified exon | in set | Reference | Length (in bp) |
Forward / reverse primer |
Name |
|---|---|---|---|---|---|
| exon 48 | a | Abbs 1991 | 506 | ttgaatacattggttaaatcccaacatg / cctgaataaagtcttccttaccacac |
ex48-F/R |
| exon 44 | b | Abbs 1991 | 426 | gttgtgtgtacatgctaggtgtgta / tccatcacccttcagaacctgatct |
ex44-F2/R |
| exon 51 | c | Abbs 1991 | 388 | gaaattggctctttagcttgtgtttc / ggagagtaaagtgattggtggaaaatc |
ex51-F/R |
| exon 43 | d | Abbs 1991 | 357 | gaacatgtcaaagtcactggacttcatgg / atatatgtgttacctacCCTTGTCGGTCC |
ex43-F/R |
| exon 45 | e | Abbs 1991 | 307 | ctttctttgccagtacaactgcatgtg / cattcctattagatctgtcgccctac |
ex45-F2/R |
| exon 50 | f | Abbs 1991 | 271 | caccaaatggattaagatgttcatgaat / tctctctcacccagtcatcacttcatag |
ex50-F/R |
| exon 53 | g | Abbs 1991 | 212 | TTGAAAGAATTCAGAATCAGTGGGATG / CTTGGTTTCTGTGATTTTCTTTTGGATTG |
ex53-F/R |
| exon 47 | h | Abbs 1991 | 181 | cgttgttgcatttgtctgtttcagTTAC / gtctaacCTTTATCCACTGGAGATTTG |
ex47-F/R |
| exon 42 | i | Abbs 1991 | 155 | CACACTGTCCGTGAAGAAACGATGATG / TTAGCACAGAGGTCAGGAGCATTGAG |
ex42-F2/R2 |
| exon 60 | j | Abbs 1991 | 139 | AGGAGAAATTGCGCCTCTGAAAGAGAACG / CTGCAGAAGCTTCCATCTGGTGTTCAGG |
ex60-F/R |
| exon 52 | k | Abbs 1991 | 113 | AATGCAGGATTTGGAACAGAGGCGTCC / TTCGATCCGTAATGATTGTTCTAGCCTC |
ex52-F/R |
1. Chamberlain JS, Gibbs RA, Ranier JE, Caskey CT (1990). Multiplex PCR for the diagnosis of Duchenne muscular dystrophy. In: PCR Protocols: A Guide to Methods and Applications (Eds; MA Innis, DH Gelfand, JJ Sninsky, TJ White), Academic Press, San Diego (USA). pp 272-281.
2. Kunkel LM, Snyder JR, Beggs AH, Boyce FM, Feener CA (1991). Searching for dystrophin gene deletions in patients with atypical presentations. In: Etiology of human diseases at the DNA level (Eds; J Lindsten, U Petterson), Raven Press, New York (USA), pp 51-60.
| Top of page |
| LMDp homepage | Remarks /
information | Copyright©,
liability |